The post 8周解析SARM1蛋白結構 appeared first on 晶泰科技 XtalPi.
]]>挑戰:
- SARM1具顯著的動態構象變化,體外難以穩定并捕捉其激活態構象;
- 目前的候選藥物分子主要為競爭性抑制劑,缺乏明確的抑制機制。
目標:
- 通過晶泰科技AI驅動的Cryo-EM圖像處理模塊,幫助加速解析抑制劑與SARM1的高分辨率電鏡結構,為理性藥物設計提供新思路、新方向;
- 進一步揭示該靶點的自抑制態與激活態的轉換機制,加深對靶點作用機理的理解。
結構解析流程:

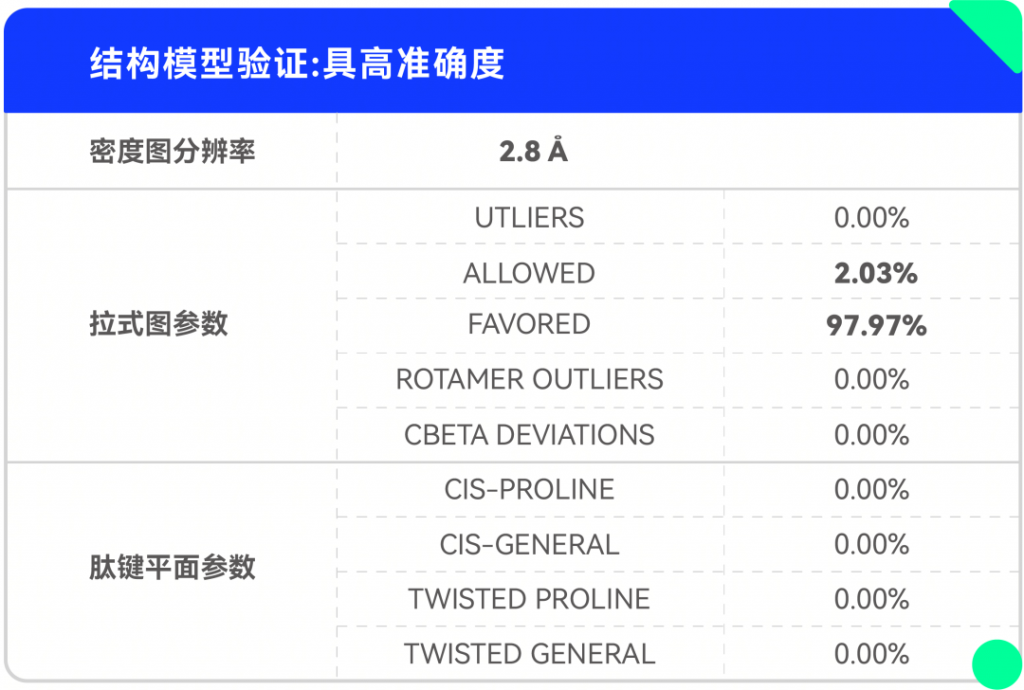
解析結果:
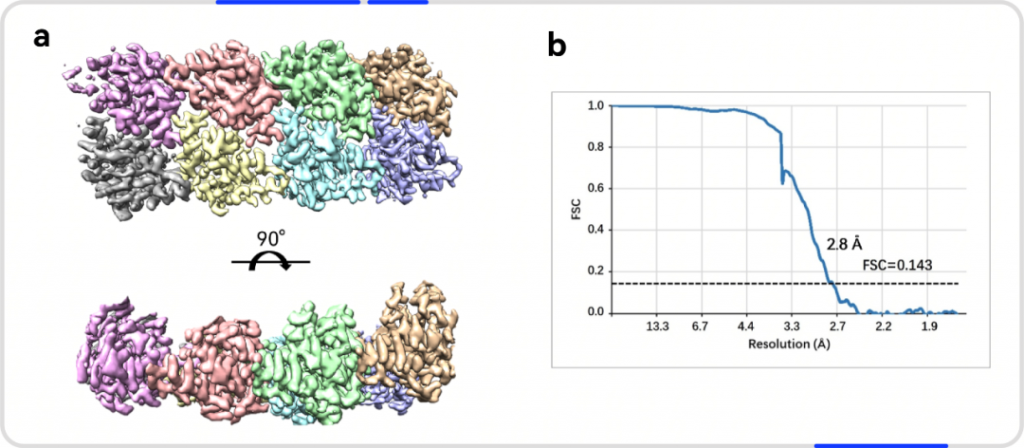
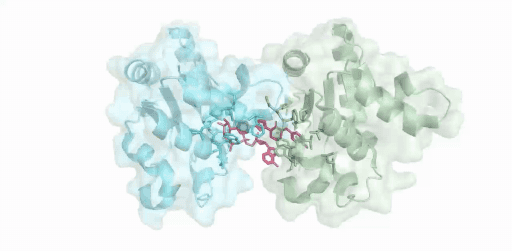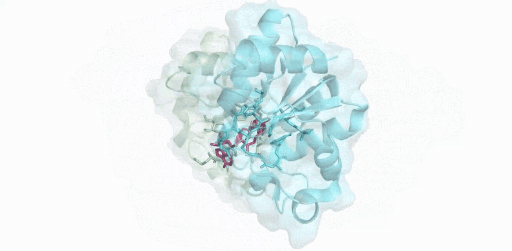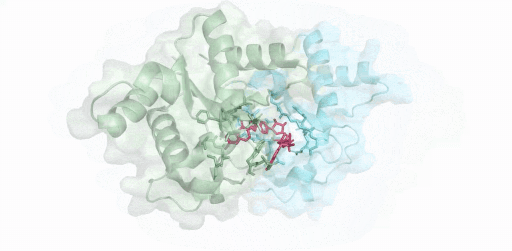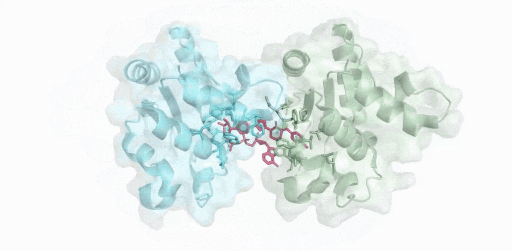
(綠色:SARM1-TIR(A),青色:SARM1-TIR(B),紅色:抑制劑-ADPR )
項目小結:
僅使用200kV電鏡,在8 周內成功解析SARM1與抑制劑的2.8 ?結構,這項開創性研究成果揭示了一種小分子抑制劑的競爭性抑制機制。高比例NMN/NAD+誘導激活SARM1,抑制劑在SARM1催化下與NAD+發生堿基交換反應,生成抑制劑-ADPR這一新產物,該產物與SARM1結合并鎖定其處于激活態構象,封鎖底物NAD+結合口袋,從而抑制SARM1的NAD+降解活性。該結果為證實SARM1的激活機制和堿基交換反應提供了最直接的結構證據。
數據計算過程中,通過使用 AI 驅動的圖像處理模塊實現了目標顆粒的精確挑選,與經典方法相比,在同等算力下,加速整體計算流程并實現更高的分辨率。
同時,首次從分子水平揭示了 SARM1 的激活機制,加深對其如何介導退行性疾病發生機理的理解。
The post 8周解析SARM1蛋白結構 appeared first on 晶泰科技 XtalPi.
]]>The post Identifying Novel FGFR3 Inhibitors By XtalPi-Curated HTS Diversity Library appeared first on 晶泰科技 XtalPi.
]]>Case Study: How XtalPi Used Diversity Library of 80K Compounds to Find Hits Targeting FGFR3
Mutations in Fibroblast Growth Factor Receptor 3 (FGFR3) are linked to various cancers, making it a critical target for therapeutic intervention. But targeting specific FGFR members individually is very challenging due to their structural similarities. Currently only pan-inhibitors are approved by the FDA.
In an effort to selectively target FGFR3, XtalPi sought to identify novel scaffolds using our curated?high-throughput screening (HTS) diversity library.
Our Key Results:
- Discovery of 15 potent hit compounds, each exhibiting biochemical potency with IC50 values below 10 μM.
- Identified three hit series which possessed novel scaffolds that are distinct from those of known FGFR3 inhibitors, providing new avenues for selectively targeting the protein.
The post Identifying Novel FGFR3 Inhibitors By XtalPi-Curated HTS Diversity Library appeared first on 晶泰科技 XtalPi.
]]>The post Hit Identification – Novel Hits for GPX4 appeared first on 晶泰科技 XtalPi.
]]>Case Study: How the XtalPi Team Discovered Novel, Non-Covalent, Potent Hits Against GPX4 in 28 Days
GPX4 is a critical enzyme for cellular antioxidant defense, but is a highly challenging target. In this case study, we share the story of how we discovered three novel non-covalent hit compounds with biochemical IC50?< 10μM in just 28 days.
- Using predictive AI models and physics-based computations, we probed key pharmacophores with high accuracy and throughput, even without reference compounds.
- 159 library compounds were proposed and 124 were successfully synthesized using our automation platform.
The post Hit Identification – Novel Hits for GPX4 appeared first on 晶泰科技 XtalPi.
]]>The post Lead Identification – Discovering Selective Inhibitors Against an Unreported Binding Pocket of SMARCA2 via a Rational Drug Design Approach appeared first on 晶泰科技 XtalPi.
]]>Case Study: How the XtalPi Team Discovered a Selective, Novel Lead Series for Key Cancer Target SMARCA2
Loss-of-function mutations in SMARCA4 occur in various cancers, leading to an increased dependency on the structurally similar SMARCA2 for activity. This makes SMARCA2 an attractive therapeutic target due to its increased importance in cancer cell survival.
However, identifying novel small-molecule SMARCA2-selective inhibitors poses significant challenges. To address this, XtalPi leveraged its advanced AI-powered discovery platform and Cryo-EM technology, opening up new possibilities for targeted cancer treatments.
Our Key Results:
- Discovered a novel lead series with high efficacy and 20-fold selectivity for SMARCA2 over SMARCA4 in biochemical assays, showcasing strong potential for further development.
- Identified a unique SMARCA2 binding pocket using Cryo-EM at 2.9 ? resolution, offering new insights for the design of selective inhibitors and advancing structure-based drug discovery.
- Achieved 100% library compound synthetic success rate by proprietary synthetic feasibility prediction models.
The post Lead Identification – Discovering Selective Inhibitors Against an Unreported Binding Pocket of SMARCA2 via a Rational Drug Design Approach appeared first on 晶泰科技 XtalPi.
]]>